Our group has completely developed from scratch a computer program for performing VS called BINDSURF, last publication reported in:
Sánchez-Linares, I., Pérez-Sánchez, H., Cecilia, J. M. & Garcia, J. M. High-Throughput parallel blind Virtual Screening using BINDSURF. BMC Bioinformatics 13, S13 (2012).
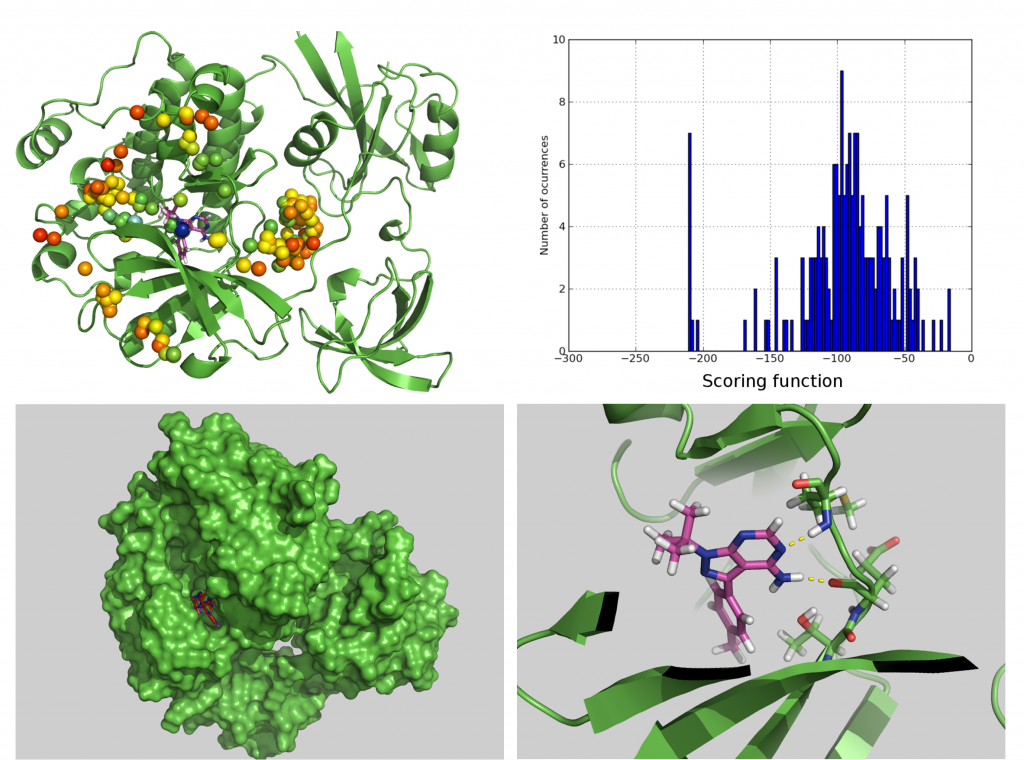
Representation of the docking results for TMI obtained over the whole surface of antithrombin. Each different simulation starts on the center of each colored bead. Color of each bead denotes scoring function value. Colors from red to blue denote stronger interactions. In order to emphasize location of binding site, crystallographic pose of heparin (pink skeleton) is shown. Predicted binding mode of TMI is shown in blue skeleton.
The VS program BINDSURF finds well-binding ligands for a protein receptor of known structure in a database that contains the three-dimensional structures of many ligands. Each ligand of the database is docked into the whole surface (see figure above) of the protein using an all-atom representation of the protein and ligand. BINDSURF does not make any assumption about the location of the binding site of the protein. After a BINDSURF execution, and with the obtained information about how different ligands dock in the protein surface, one can start to make hypothesis about different potential binding sites that can guide posterior and more detailed analysis using other VS methods. BINDSURF is an efficient and fast methodology for the unbiased determination on GPUs of protein binding sites depending on the ligand. It can also be used for fast pre-screening of a large ligand database, and its results can guide posterior detailed application of other VS methods (such as Molecular Dynamics and multiscale simulation methods).
Our program BINDSURF provides in a short time frame accurate and reproducible ligand binding poses for a great number of experimentally known complexes. To the best of our knowledge, there is no other method that can perform whole protein surface screening in these conditions and at such speed. This drastically facilitates screening of databases containing up to milllions of chemical compounds thus increasing chances of bioactive molecule discovery.